Artificial intelligence to develop drugs in record time

When a machine learning algorithm is used to predict which diseases a drug can be used to treat, it is not possible to explain why, and this raises doubts about the reliability of the prediction. Therefore, it would be ideal to have a repositioning mechanism that also explains why it makes the predictions it does.
This is where a group of researchers from the Universidad Politécnica de Madrid (UPM) has just developed a drug repositioning method with a clear emphasis on interpretability, as it seeks to explain why it proposes treating disease X with drug Y. This new algorithm, called ‘XG4REPO’ (eXplainable Graphs for Repurposing), not only repurposes drugs but also presents the results in an understandable way, indicating which biological mechanisms are used for the prediction. This allows its predictions to be validated by medical experts, who can assess whether it is a valid explanation or not, thus generating much more robust predictions.

Own creation with images from Pixabay
The process of creating medicines is slow and costly, as it involves numerous tests to ensure the safety of the new medicine in order to obtain marketing authorisation from the health authorities. An alternative that is gaining momentum to alleviate this situation is the repositioning of existing medicines, which consists of identifying new applications for drugs that are already approved. This means using an existing drug to treat a disease other than the one for which it was originally designed.
The repositioning of drugs, as a technique, has a number of advantages that cannot be ignored. The first is that it significantly shortens drug development times, as the drug is already approved and its side effects are known. The second major advantage relates to the cost of drug development, as there is no need to repeat costly safety tests, first on animals and then in clinical trials. Furthermore, most drugs developed in laboratories are never marketed due to their adverse effects, a problem that repositioning does not have.
These advantages mean that repurposing is considered a technique that can bring about major changes in medicine. On the one hand, it allows treatments for new diseases to be developed much more quickly than creating a drug from scratch. During the COVID pandemic, for example, repurposing made headlines due to attempts to use different drugs to treat this new disease. But repurposing also offers great hope for patients with rare diseases, as it would allow the development of treatments at a low cost to the laboratory.
On a technical level, a medicine affects a specific biological process; for example, paracetamol blocks part of the pain impulse in the human body. The great challenge of repositioning is to identify which patterns, affected by a specific medicine, appear in other diseases. Thus, if two diseases have a similar pattern, and a certain medicine is used to treat the first, it is likely that it can also be used for the second. However, identifying these patterns by experts is a costly process that requires extensive knowledge of diseases and their mechanisms. Nevertheless, the machine learning techniques currently available are very good at detecting patterns, hence the recent focus on drug repositioning using artificial intelligence.
However, artificial intelligence techniques present the problem of interpretability. To resolve this, a group of researchers from the UPM, Escuela Técnica Superior de Ingenieros de Telecomunicación (ETSIT) has found the solution to this problem thanks to the design of a new algorithm they have called ‘XG4Repo’, which provides a framework for drug repurposing using knowledge graphs that predict diseases that can be treated with a given compound. Along with the prediction, the model provides the rules that support the prediction and the importance of the rule.
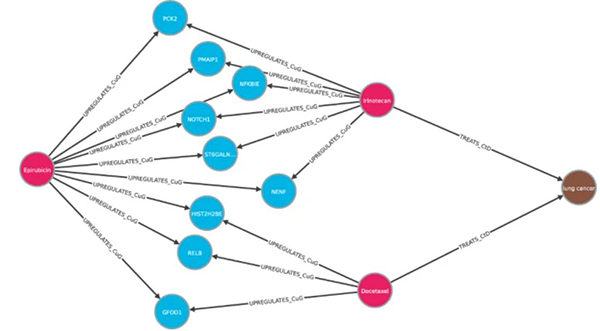
Example of explainable repositioning: the biological pathways that XG4REPO provides as justification for treating lung cancer with Epirubicin are indicated. Source: Jiménez, A., Merino, M.J., Parras, J. et al. Explainable drug repurposing via path-based knowledge graph completion. Scientific Report 14.
To demonstrate the effectiveness of ‘XG4REPO’, researchers tested the algorithm’s ability to predict the application of three known cancer drugs and found that many of the algorithm’s predictions were already in the initial clinical trial phase. This means that there is medical evidence to validate the predictions made by ‘XG4REPO’. Therefore, as Professor Santiago Zazo, who was part of the working team, points out, ‘this mechanism constitutes another step towards the application of artificial intelligence techniques in the medical field, not to replace experts, but to facilitate the analysis of large amounts of data in a short time and accelerate the drug development process’.
Bibliographic reference:: Jiménez, A., Merino, M.J., Parras, J. et al. Explainable drug repurposing via path based knowledge graph completion. Sci Rep 14, 16587 (2024).
https://doi.org/10.1038/s41598-024-67163-x
Source: UPM, Press room, Research news: https://short.upm.es/kroq0, publicado en su fuente original el 21.10.2024
Royalty-free cover image:Imagen de freepik
Share this:
Latest news



Categories

